SemanticSBML
Documentation of the Merge Algorithms
This is the documentation of the annotation algorithms used in semanticSBML. Here you can read about the following topics:
Main Idea
To enable a merging of models that will result in a biological meaning full result, semanticSBML uses an abstraction of a model that is different from those of SBML. It is based on the following concept:
A
systems biology model consists of multiple biological entities. Biological entities can be identified.
ATP is KEGG#C00002 (Adenosine
5'-triphosphate)
Each
entity has a biological quantity that describes the
entity.
ATP
in the cytosol measured in mole
A model makes statements about biological entities
ATP
has the initial amount of 5 (mole)
Concept of the Merge Algorithm
Next to the model abstraction the the mergin process in semanticSBML uses further concepts that will be introduced in this section. In the following class names (written in typewriter font) are used along with artificial names for datastructures. LibSBML elements are written in italic font.
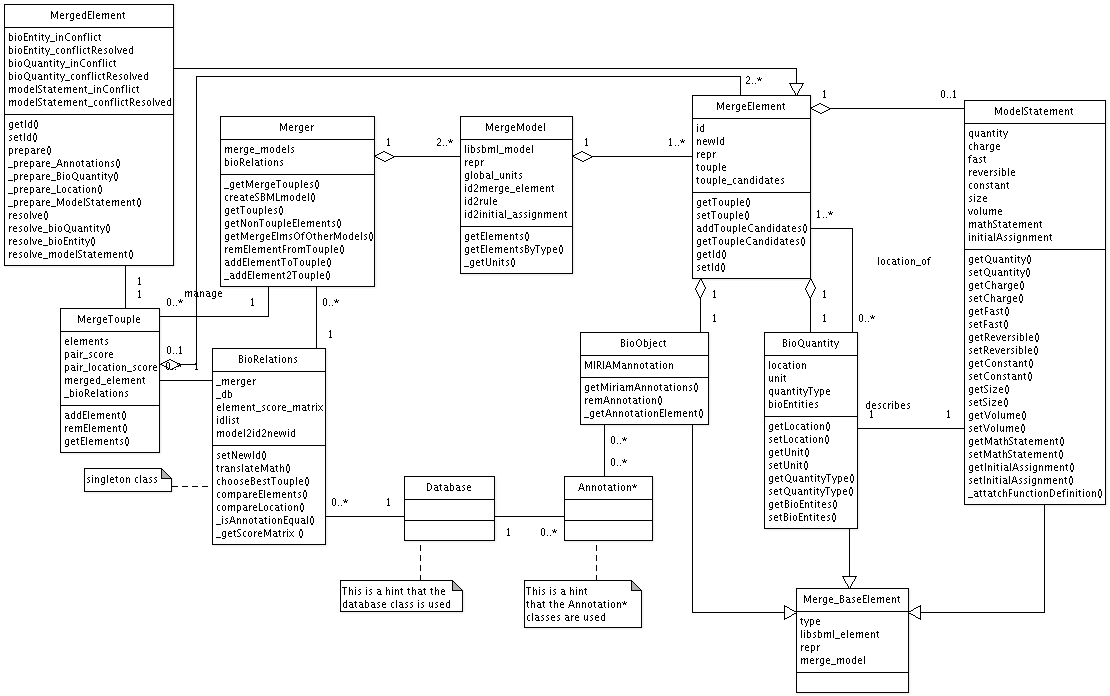
The first step is the translation of a SBML model to a semanticSBML model (see Figure merge_cartoon0 step 1; the steps are marked with large grey numbers). On this account the libSBML base elements (see Appendix ???) compartment, species, parameter, reaction (these elements are considered as mergable biological entities) are translated into MergeEntitys. The MergeEntity class can be viewed as meta SBML element since it contains all of the attributes the different types of SBML elements can contain. In some cases an attribute can also be a SBML base element. The MergeModel stores all MergeEntitys of one libSBML model. and provides access to libSBML elements.
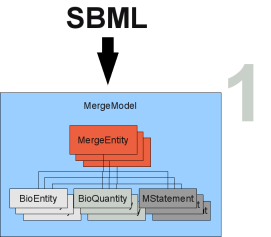
 The translation is repeated for all models that should be merged (see Figure merge_cartoon1 step 2). All elements in each model are compared pairwise with the elements (of matching type) in the other models (see Figure merge_cartoon1 step 3). The algorithm checks for biological identity with the help of MIRIAM annotations as well as structural identity (location of physical biological entities e.g., ATP in cytosol - ATP in mitochondrion).
The translation is repeated for all models that should be merged (see Figure merge_cartoon1 step 2). All elements in each model are compared pairwise with the elements (of matching type) in the other models (see Figure merge_cartoon1 step 3). The algorithm checks for biological identity with the help of MIRIAM annotations as well as structural identity (location of physical biological entities e.g., ATP in cytosol - ATP in mitochondrion).
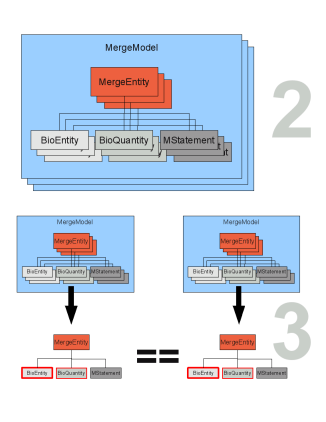
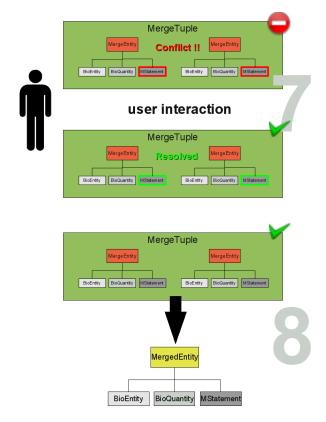
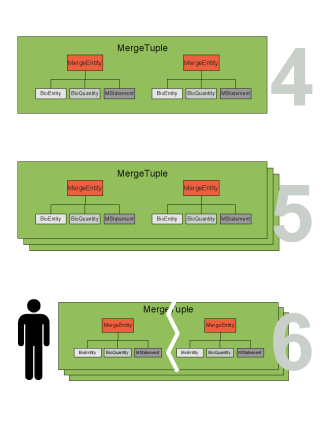 If duplicate entities are found they are stored in a MergeTuple (see Figure merge_cartoon2 step 4). The MergeTuple is a smart container that only allows the storage of one entity from each model (an aggregation of entities is not allowed). It also decides which element belongs in the container. A list of MergeTuples is generated (see Figure merge_cartoon2 step 5). The user can manually modify this list since a correct matching of the duplicate entities can not be guaranteed (see Figure merge_cartoon2 step 6). Even though it is not desired, this also enables the manual merging of non annotated models.
If duplicate entities are found they are stored in a MergeTuple (see Figure merge_cartoon2 step 4). The MergeTuple is a smart container that only allows the storage of one entity from each model (an aggregation of entities is not allowed). It also decides which element belongs in the container. A list of MergeTuples is generated (see Figure merge_cartoon2 step 5). The user can manually modify this list since a correct matching of the duplicate entities can not be guaranteed (see Figure merge_cartoon2 step 6). Even though it is not desired, this also enables the manual merging of non annotated models.
The entities in the MergeTuple may contain conflicting values. Most conflicting values can be resolved by choosing a value from a list e.g., choosing the correct initial amount of the ATP (see Figure merge_cartoon3 step 7). However conflicts in the BioEntiy (determining the identity of the entity) or the BioQuantiy (describing the entity) show that there is a severe disagreement between the entities. The resolution of severe conflicts may need more than a simple choice and should often lead to the destruction of the tuple. The user can however solve every conflict and thus mark the MergeTuple as resolved. From a resolved or non conflicting MergeTuple a combined element is created called the MergedEntity (see Figure merge_cartoon3 step 8). The MergedEntity has the same properties as a MergeEntity and has only implementational importance.
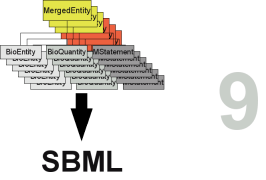
 From the list of all MergedEntitys and MergeEntitys a merged SBML model can be created (see Figure merge_cartoon2 step 8). The merged SBML model integrates all information of the inserted models.
From the list of all MergedEntitys and MergeEntitys a merged SBML model can be created (see Figure merge_cartoon2 step 8). The merged SBML model integrates all information of the inserted models.
The semanticSBML merge algorithm tries to realize this concept of the merge process however some changes had to be made for an enhanced usability.
UML Class Diagram

 The translation is repeated for all models that should be merged (see Figure merge_cartoon1 step 2). All elements in each model are compared pairwise with the elements (of matching type) in the other models (see Figure merge_cartoon1 step 3). The algorithm checks for biological identity with the help of MIRIAM annotations as well as structural identity (location of physical biological entities e.g., ATP in cytosol - ATP in mitochondrion).
The translation is repeated for all models that should be merged (see Figure merge_cartoon1 step 2). All elements in each model are compared pairwise with the elements (of matching type) in the other models (see Figure merge_cartoon1 step 3). The algorithm checks for biological identity with the help of MIRIAM annotations as well as structural identity (location of physical biological entities e.g., ATP in cytosol - ATP in mitochondrion). If duplicate entities are found they are stored in a MergeTuple (see Figure merge_cartoon2 step 4). The MergeTuple is a smart container that only allows the storage of one entity from each model (an aggregation of entities is not allowed). It also decides which element belongs in the container. A list of MergeTuples is generated (see Figure merge_cartoon2 step 5). The user can manually modify this list since a correct matching of the duplicate entities can not be guaranteed (see Figure merge_cartoon2 step 6). Even though it is not desired, this also enables the manual merging of non annotated models.
If duplicate entities are found they are stored in a MergeTuple (see Figure merge_cartoon2 step 4). The MergeTuple is a smart container that only allows the storage of one entity from each model (an aggregation of entities is not allowed). It also decides which element belongs in the container. A list of MergeTuples is generated (see Figure merge_cartoon2 step 5). The user can manually modify this list since a correct matching of the duplicate entities can not be guaranteed (see Figure merge_cartoon2 step 6). Even though it is not desired, this also enables the manual merging of non annotated models. From the list of all MergedEntitys and MergeEntitys a merged SBML model can be created (see Figure merge_cartoon2 step 8). The merged SBML model integrates all information of the inserted models.
From the list of all MergedEntitys and MergeEntitys a merged SBML model can be created (see Figure merge_cartoon2 step 8). The merged SBML model integrates all information of the inserted models.