This is the documentation of the annotation algorithms used in semanticSBML. Here you can read about the following topics:
urn:miriam:database.subdatabase:identifier. The
predicate describes the relationship between the subject and the
object. It is given by BioModels.net qualifier elements. The following
example shows a MIRIAM annotation in SBMtL (the example is only part of a larger
SBML document "..." denotes shortened parts).The
example above shows the SBML element species. The element is annotated
with a reference to the KEGG database identifier for Phosphorus. The
MIRIAM annotation in human words states "The element species (ATP) has a part that is the identifier C06262 (Phosphorus) of the database http://www.genome.jp/kegg/ (KEGG)". The table below explains
the important sections of the example.
| Example: | ATP | hasPart | KEGG | # | C06262 (Phosphorus) |
| RDF: | Subject | Predicate | Object |
|
|
| SBML: | Species Reaction Compartment Model ... |
BioModels.net Qualifier |
Resource |
|
|
| semanticSBML: | Entity | Qualifier | Database | # | ID |
The current version of semanticSBML supports reading the URI and URN scheme to link to external resources. However it will only write the URN scheme!
The main concept of the annotation algorithm is that is a
simplified abstraction of a SBML model (see Figure below). The main
idea is that a model consists of elements that can be annotated. The
type of an element is defined by an attribute and not by the element
itself like in SBML.
The
semanticSBML annotation concept is as follows: a model consists of
a collection of elements and elements consist of a collection of
annotations. Thus the model depends on its elements and elements
depend on its annotations. However an annotation can be used
independently from an element and an element can be used
independently from a model. The construction of the model and element
depend on the SBML whereas the annotation can be created
independently from SBML. The functions of each class will be explained
in the next section. An UML diagram of the annotation algorithm can be
found here.

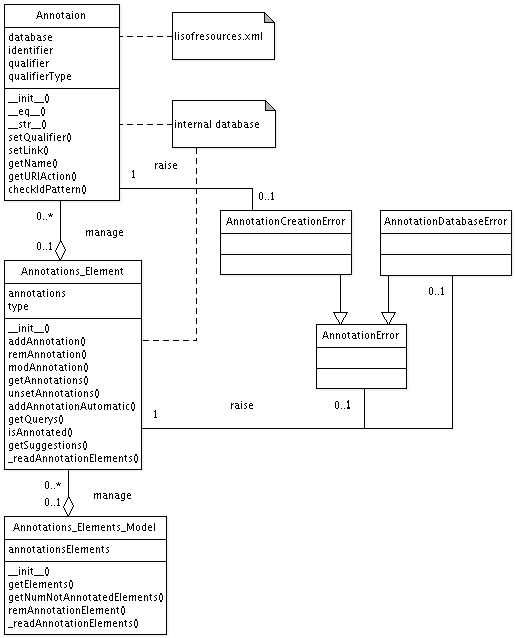
The Annotation class represents a single MIRIAM annotation. It
consists of methods to get and set the variables: database,
identifier, qualifier, qualifier type. When setting these variables
multiple checks are executed to verify their correctness. The class
uses the external file listofresources.xml.
In addition the class provides functions for comparison of
annotations and different representations of the
annotation.
Function
Description
__init__
Set
the resource (database and identifier) and qualifier (qualifier and
qual-
ifier type).
__eq__
Equality operator. If database and identifier are the same return
True.
__str__
Return the
annotation resource as specified by the proposed MIRIAM
annotation standard: URI#ID.
getName
Return a human readable string representation of the annotation - if
the
resource can be found in the
internal database or an empty string if the
resource can not be found.
getURIAction
If possible return a hyperlink to find the referenced element on the
world
wide web. This function is
dependent on the resource listofresources.
setQualifier
Set the qualifier of the annotation - libSBML encoded the
biological-
qualifiers in numbers
between 0 and 7 and model-qualifiers between 0
and 2 and qualifier-types between 0 and 2. Both numbers and string
rep-
resentations (e.g., hasPart)
are allowed as input for the qualifier and the
qualifier type. If the qualifier is not recognized an error is
raised.
setLink
Set the
database and the identifier of the Annotation class. As input
for
the database a URI (e.g.,
http://www.geneontology.org) or name (e.g.,
Gene Ontology) are both accepted. The input allows setting a flag
upon
which the insertion of unknown
databases (known ones are specified in
listofresources.xml) raise an error. If the identifier pattern is
known
for the inserted database and
the inserted identifier does not match this
pattern an error is raised (see checkIdPattern). This function is
dependent
on the resource listofresources.
checkIdPattern
This function checks if a regular expression pattern for identifiers
of a given
database can be found
(listofresources.xml). If a pattern is found the
inserted identifier is matched against the pattern. If the pattern
matches
the function returns True,
if it does not match the function returns False.
This function is dependent on the resource listofresources.
The Annotations Element class represents one annotatable
object in a model (a biological entity / libSBML base element e.g.
species, reaction, compartment). It is a container for Annotation
class instances. It has functions to add, remove and modify
annotations.
Function
Description
__init__
Read MIRIAM annotations, type, id, metaid and name from
libSBML
element. Only MIRIAM
annotatable libSBML elements are allowed as
input.
_readAnnotations
Internally used function to read all MIRIAM annotations (from
CVTerms)
of the inserted
libSBML element
isAnnotated
Return if the element contains MIRIAM
annotations.
addAnnotation
Add a MIRIAM annotation to the SBML element represented by this
class
instance. In libSBML
versions <3.0.2 the adding of identical annotations
will create (two) separate identifiers. As a result of this work
libSBML
prevents this in
later version. (The function also checked if a CVTerm with
the same qualifier already exists and add annotation to this CVTerm -
this
functionality was
discontinued since libSBML already provides it.)
modAnnotation
Modify the qualifier of an annotation.
remAnnotation
Delete an annotation from libSBML element and resynchronized the
in-
ternal list of
annotations with libSBML element.
unsetAnnotations
Delete all annotations of this element. This function is not present
in the
current libSBML but
might be added in future versions. It was created
due to the behavior of libSBML <3.0.2 described in
addAnnotation.
getAnnotations
Return a list of Annotation class instances. These instances should
be
used to add or remove
annotations.
getQuerys
Return a list of the name, id and metaid value of the libSBML
element.
These can be used to
query annotations from the internal database.
getSuggestions
Get a list of Annotations by querying the internal database. If no
query
is specified as input
the function getQuerys is used. A switch disables the
fuzzy database search, then only exact matches are
returned.
addAnnotationAutomatic
Check if the element is already annotated (using isAnnotated). If is
not
annotated, check if an
annotation can be found using getSuggestions
(exact matches only). Add annotation(s) that were found to this
element
(using addAnnotation).
The Annotations Elements Model represents a complete model. It is
a container for Annotations Element instances. Its functions are
limited since it is mainly used as a data container.
Function
Description
__init__
Read
all elements from a SBML model that can be annotated and create
a list of Annotations Elements (using
readAnnotationElements).
getNumNotAnnotatedElements
Return the number of elements that have no MIRIAM
annotations.
getAnnotationElements
Return the list of Annotations Element (that can contain MIRIAM
an-
notations) available in this
model.
remAnnotationElement
Remove an element.
readAnnotationElements
Go through all MIRIAM annotatable elements in a libSBML model
and
create Annotations
Elements.
In addition to these classes the Merger class of
SBMLmerge was replicated for backwards compatibility. It is a simple
wrapper class. Most of its functions can be found (with different
names) in one of the classes above.
The following example shows the usage of the model class. It is initialized with a libSBML Model instance. AnnotaionErrors have to be caught in case the Model has elements with faulty annotations.
1 import libsbml
2 from semanticSBML.annotate import *
4 document=libsbml.readSBMLFromString(open("mymodel.xml","r").read())
5 try:
6 aem=Annotations_Elements_Model(document.getModel())
7 except AnnotationError,e:
8 print e
9 else:
10 print aem.getNumNotAnnotatedElements()
The Annotations Element class is initialized with a libSBML element in this case a species element. Like in the initialization of the model the element might raise an exception if the annotations of the element are faulty. On a successful initialization the number of non annotated models is printed to the screen in the example below.
11 try:
12 ae=Annotations_Element(list(document.getModel().getListOfSpecies())[0])
13 except AnnotationError,e:
14 print e
15 else:
16 print ae.isAnnotated()
Annotation instance can be created in different ways. In line 17 and 18 identical annotations are created using different input. In line 17 human readable representations are used in line 18 the URI and the libSBML numbers of the BioModels qualifiers are used (see Section 3.2.4). Line 19 creates an annotation for a model.
17 a1=Annotation("Gene Ontology","GO:1234567","bio","is")
18 a2=Annotation("http://www.geneontology.org/","GO:1234567","1","0")
19 a3=Annotation("BioModels","BIOMD0000000001","model","is")
The annotation objects created in the example above can be used to add annotations to a model.
20 ae.addAnnotation(a1)
The adding of the annotation will modify the libSBML model instance.
To save the changes persistently the model has to be written to a
file on the hard disk by using libSBML functions.
Annotation suggestions and automated annotation rely on similar features. To automatically find annotation suggestions the properties name, id and metaid of an SBML element are collected (function getQuerys Annotations_Element Class) and used for a search the internal database of semanticSBML. The database currently has two search features: exact search, fuzzy search. The Annotations_Element class wraps these features in the function getSuggestions. The function has two optional inputs: query (if empty use getQuerys) and exact_search (default False). In an autmated annotation the function getSuggestions is called for every element. The returned Annotations are added to the SBML element. This means that an exact search, returning only database entries that exactly match the search term, is executed with the results of getQuerys. The fuzzy search is used when a user searches for a specific term. For a fuzzy search the python search function difflib.get_close_matches is used.