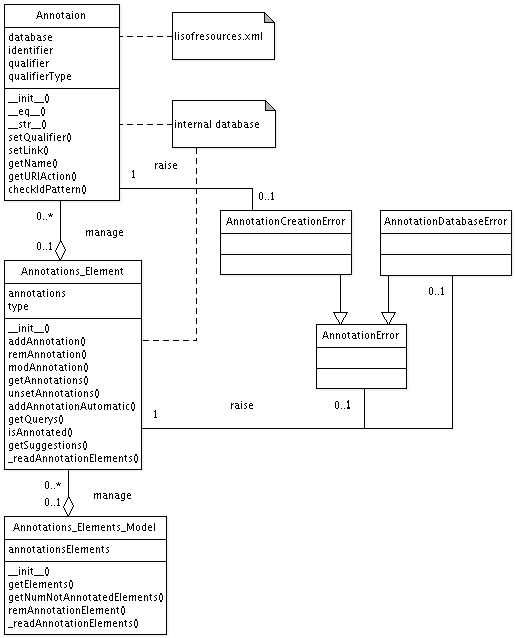
An XML file was created that contains all known databases that can be used for MIRIAM annotations. The XML file is based on an XML file created by the BioModels Database team of Nicolas Le Novère. This is an example entry
<resource name="EC code"Meaning of the variables
uri="http://www.ebi.ac.uk/IntEnz/"
alternateUris='http://www.ec-code.org/'
location="http://www.ebi.ac.uk/IntEnz/"
action="http://www.ebi.ac.uk/intenz/query?cmd=SearchEC&ec="
elements="assignmentRule rateRule algebraicRule reaction event"
idInputHelper=""
idPattern="^(\d+|\d+\.(-|\d+)|\d+\.\d+\.(-|\d+)|\d+\.\d+\.\d+\.(-|\d+))$"
/>
| name | Database Name |
| uri | MIRIAM resource uri part |
| alternateUris | alternate Uris of MIRIAM resources that have been found |
| location | World Wide Web location |
| action | URL that can be called in combination with the Database Identifier that will show a web page with the biological object |
| elements | space separated list of libSBML elements that can be annotated with this database |
| idPattern | regular expression pattern that the database id must match |
The XML file libSBML resources contains the default unit definitions as specified in the libSBML specifications, all known units, as well as the current BioModels.net qualifiers, with descriptions and mappings to the Ids used in libSBML. The file can be downloaded from the SVN repository.
The automatic matching of libSBML elements in the merging process is done with the help of a rather simple algorithm. The algorithm in pseudo code can be seen below
function compare_SBMLelements(entity1,entity2):
score=0
for (an1=MIRIAM annotation) in entity1:
for (an2=MIRIAM annotation) in entity2:
if is_equal(an1, an2):
score+=score_matrix[an1.qualifier][ an2.qualifier]
return score
function is_equal(an1,an2):
if an1==an2: return True
elseif database.is_equal(an1,an2) : return True
else:
The
score matrix (score_matrix[an1.qualifier][ an2.qualifier]) which is
also located in the .semanticSBML directory currently looks as follows.
is hasPart isPartOf isVersionOf hasVersion isHomologTo isDescribedBy unknown
is 10 1 1 1 1 1 1 1
hasPart 1 1 1 1 1 1 1
isPartOf 1 1 1 1 1 1
isVersionOf 1 1 1 1 1
hasVersion 1 1 1 1
isHomologTo 1 1 1
isDescribedBy 1 1
unknown 1
Even though the algorithm is very naive the largest problem is to improve the function is_equal(). The matching of elements that have e.g. a parent-child relationship (e.g. Glucose and α-D-glucose) is currently not possible.
