Introduction
SBML2LaTeX converts files in the System Biology Markup Language (SBML) format into LaTeX files. A convenient online version is available, which allows the user to directly generate various file types from SBML including PDF, TeX, DVI, PS, EPS, GIF, JPG or PNG. SBML2LaTeX can also be downloaded and used locally in batch mode or interactively with its Graphical User Interface or several command line options. The purpose of SBML2LaTeX is to provide a way to read the contents of XML-based SBML files. This is helpful and important for, e.g., error detection, proofreading and model communication.
Please cite: Dräger A, Planatscher H, Wouamba DM, Schröder A, Hucka M, Endler L, Golebiewski M, Müller W, and Zell A: “SBML2LaTeX: Conversion of SBML files into human-readable reports”, Bioinformatics 2009. [BibTeX], [doi:10.1093/bioinformatics/btp170] (draeger - 2009-03-29 19:29)
Availability of SBML2LaTeX
SBML2LaTeX is available in two different forms:
- As a stand-alone application that converts SBML files into LaTeX files. This application also includes a convenient script displaySBML.sh, which allows further processing of the LaTeX files.
- As a online version, which allows users to directly create various human-readable files including PDF, DVI and PS without installation of any software on the local computer.
The online version
This convenient online version allows direct creation of SBML model reports from a given SBML file without local installation of any software. Several output formats are available, the font size and style, paper size, and page orientation can be changed. Landscape pages are very useful because it is often not possible to introduce a line break into long denominators and the resulting formula therefore does not fit on portrait pages. The following features can be switched off: translation of MIRIAM annotations to online resources, SBML consistency check, and the inclusion of all predefined units. Other layout options enable the user to choose whether the details of all reaction participants should be listed in one table or separate tables for each group (reactants, modifiers and products). Instead of a simple headline, a separate title page can be created.
The stand-alone version
The stand-alone tool provides a Graphical User Interface (GUI) mode and a command-line mode. All options (see section options) to customize the output (layout, SBML properties to be included) are available through both modes. This allows users to use SBML2LaTeX in a batch mode or with user interaction. The GUI can be started by passing the --gui option to the program. All other options passed to the program are adopted by the GUI.
An API documentation can be found at here, which allows users to costumize SBML2LaTeX or to access its methods from their own programs.
To process the results of this stand-alone program to obtain, e. g., PDF files, an installation of LaTeX and several additional packages on your system is required. For details see the LaTeX section.
A convenient Bash script is available on the download section, which enables the user to directly create PDF, DVI, PS, EPS, JPG, GIF or TeX files. This script can also be used in combination with the graphical user interface. For details see section displaySBML.sh.
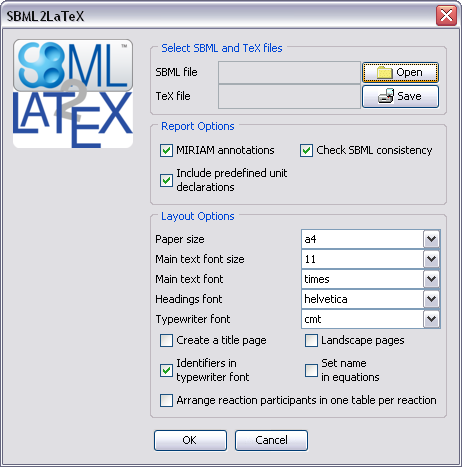
Command line options
java [-cp <classpath definition>] SBMLconverter [in_file.xml] [out_file.tex] [options]
Note that it is probably necessary to set the environment variable LD_LIBRARY_PATH to the location of libSBML together with its XML parser library before calling SBMLconverter.
Usage of the script displaySBML.sh
displaySBML.sh in_file.xml [tex|latex|pdf|ps|eps|dvi|jpg|png|gif] [options]
This script accepts the same options as SBMLconverter itself. Note that it is, however, probably necessary to customize this shell script because some paths within the script may not point to the correct locations on your system. It is also important to know that this script requires LaTeX, ImageMagick and ZIP to be installed on your system in addition to all requirements of SBML2LaTeX itself. For details see the download section.
Possible command line options (case and ordering
are ignored)
1. Options to include or exclude SBML information
| --show-predefined-units=<true|false> | If true (default), all predefined unit definitions of SBML are made explicit. |
| --miriam=<true|false> | If true (default), MIRIAM annotations are included into the model report if there are any. This option may require the path to the MIRIAM translation file to be specified. |
| --check-consistency=<true|false> | |
| If true, the libSBML model consistency check is performed and the results are written in the glossary of the model report. | |
2. Layout options for the model report
| --paper-size=<a<0..9>|b<0..9>|c<0..9>|d<0..9>|letter|legal|executive> | With this option the paper format can be influenced. Default paper size: DIN A4. All sizes a?, b?, c? and d? are European DIN sizes. Letter, legal and executive are US paper formats. |
| --landscape=<true|false> | If true, the whole report is written on landscape paper instead of portrait format. |
| --title-page=<true|false> | If true, an extra title page will be created. |
| --font-size=<8|9|10|11|12|14|17> | This option allows you to select a smaller or larger font (for continuous text). Default size: 11 pt. |
| --font-text=<chancery|charter|cmr|palatino|times|utopia> | Allows to select the font of continuous text. Default: times. |
| --font-headings=<avant|cmss|helvetica> | Allows to select the font of captions and other sans serif text. Default: helvetica |
| --typewriter=<true|false> | If true (default) all identifiers are written in typewriter font. |
| --font-typewriter=<cmt|courier> | Selects the font to be used for typewriter text. Default: cmt. |
| --print-name-if-available=<true|false> | If true, the names of elements are used in equations instead of their identifiers. |
| --reactants-overview-table=<true|false> | If true, the details (identifier and name) of all reactants, modifiers and products participating in a reaction are listed in one table. By default a separate table is created for each one of the three participant groups including its SBO term. |
3. Definition of required file paths
| --logo-file=<path to logo file (a graphics file)> | The SBML logo to be displayed at the beginning of the model report. Default path: resources/SBML2LaTeX.pdf |
| --sbo-file=<path to SBO obo file> | The file containing the SBO translation in OBO format. Default path: resources/SBO_OBO.obo |
| --miriam-file=<path to MIRIAM XML file> | The file containing the translation of URNs/URIs to actual URLs. Default path: resources/MIRIAM.xml |
| --html-config-file=<path to HTML2LaTeX configuration file> | The configuration file for the translation of notes in XHTML format to LaTeX commands and text. Default path: resources/config.xml |
4. General program options
| --gui | Opens a dialog window for convenient usage.
With this option no in/out-file needs to be
specified. All other command line options are
accepted and their values influence the
behavior of the dialog window.
See the following
screenshot |
| --help | Shows this overview. |
If the resulst of SBML2LaTeX are to be translated to DVI, PS or PDF files, an installation of LaTeX is required. In addition to standard LaTeX, SBML2LaTeX uses several packages, which you will need to install to your system or to copy them into the same directory as the results of SBML2LaTeX. Another option is to copy the following style packages into a subdirectory of your SBML2LaTeX installation and to set the TEXINPUTS variable to this directory (see the displaySBML.sh script for an example). For convenience, all packages listed below are already included in the SBML2LaTeX bundle, besides the KOMAscript. Required LaTeX styles and packages:
- KOMAscri pt (should already be included into your LaTeX installation)
- breqn introduces automatic line breaks within equations.
- flexisym is needed by breqn.
- mhchem required to typeset chemical equations.
- rccol produces fancy tables in which decimal numbers always get the same number of decimal digits.
- fltpoint is required by rccol.
The software license of SBML2LaTeXThis program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version. |

|
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/gpl.html.
Included third-party libraries and packages
- SBML2LaTeX uses and includes software developed by the JDOM Project (http://www.jdom.org), which is available under the appache license.
- SBML2LaTeX uses and includes HTML2LaTeX (see http://htmltolatex.sourceforge.net), which is freely available under the GPL version 2 license.
- SBML2LaTeX delivers several unmodified LaTeX packages for the sake of simplicity. These are all freely available under the LaTeX Project Public License.
Acknowledgments
We are greatful to everybody, who participated in this work, especially Jochen Supper, Henning Schmidt, Detlev Bannasch, the SBML team (see http://sbml.org/SBML.org:About), and the LaTeX3 team, especially the developer of the breqn package that introduces automatic line breaks into equations, Morten Høgholm.
This project was funded by the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF) in the projects National Genome Research Network (Nationales Genomforschungsnetz, NGFN-Plus) and HepatoSys “Kompetenznetzwerk Systembiologie des Hepatozyten”, and supported by the Center for Bioinformatics Tübingen (Zentrum für Bioinformatik Tübingen, ZBIT). The German federal state Baden-Württenberg funded the Tübinger Bioinformatik-Grid project that enabled us to provide the convenient SBML2LaTeX web service.
SBML2LaTeX includes software developed by the JDOM Project (http://www.jdom.org).
We are grateful to the developers of HTML2LaTeX because for the conversion of XHTML elements within the SBML notes element, SBML2LaTeX includes a modified version of this program (see http://htmltolatex.sourceforge.net).

